A comparative study of filter methods based on information entropy
Copyright © The Korean Society of Marine Engineering
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0), which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Feature selection has become an essential technique to reduce the dimensionality of data sets. Many features are frequently irrelevant or redundant for the classification tasks. The purpose of feature selection is to select relevant features and remove irrelevant and redundant features. Applications of the feature selection range from text processing, face recognition, bioinformatics, speaker verification, and medical diagnosis to financial domains. In this study, we focus on filter methods based on information entropy : IG (Information Gain), FCBF (Fast Correlation Based Filter), and mRMR (minimum Redundancy Maximum Relevance). FCBF has the advantage of reducing computational burden by eliminating the redundant features that satisfy the condition of approximate Markov blanket. However, FCBF considers only the relevance between the feature and the class in order to select the best features, thus failing to take into consideration the interaction between features. In this paper, we propose an improved FCBF to overcome this shortcoming. We also perform a comparative study to evaluate the performance of the proposed method.
Keywords:
Metaheuristics, Improved tabu search, Subset selection problem1. Introduction
Since the advent of big data, feature selection has played a major role in reducing the “high-dimensionality”. Feature selection improves the performance of machine learning algorithms and helps to overcome the limited storage requirements, and ultimately reduces costs. Feature selection is to select relevant features and remove irrelevant and redundant features. It has been widely employed in applications ranging from text processing, face recognition, bioinformatics, speaker verification,and medical diagnosis to financial domains.
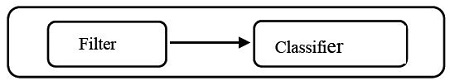
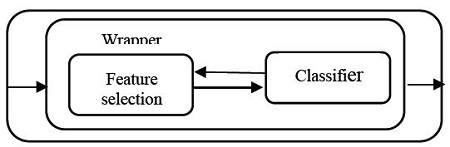
Feature selection methods can usually be classified into four categories : filter [1][2], embedded [3][4], wrapper [5]-[7], and hybrid methods [8]-[10]. Filter methods (see Figure 1) use variable ranking techniques without considering any learning classifier such as SVM (support vector machine) [11], NB (naïve Bayesian) [12][13], kNN (k-nearest neighbor) [14], and DT (decision tree) [15][16]. Unlike the filter methods, wrapper methods (see Figure 2) select a feature subset using a learning classifier as part of the evaluation function.
Filter methods can be broadly divided into two classes: univariate and multivariate approaches. Univariate approaches evaluate the relevance of each feature individually, and then select a subset of features having the highest ranks. Several univariate criteria have been developed in the literature including GI (gini index) [17], IG (information gain) [18] , Chi-square test [19], FS (Fisher score) [20], LS (Laplacian score) [21], and Relief [22]. The univaiate filter methods are computationally very efficient due to the ignorance of the dependency between features. Thus, with univariate approaches, computing time is extremely fast, but they produce less accurate solutions.
To overcome this flaw of the univariate filter, in which the dependency between features is ignored, multivariate approaches have been proposed in the literature. The FCBF (Fast Correlation-Based Filter) [23] and mRMR (minimum Redundancy Maximum Relevance) [24] are well-known efficient multivariate approaches.
In this paper, we focus on filter methods based on information entropy : IG, FCBF, and mRMR. FCBF has the advantage of reducing computational burden by eliminating the redundant features that satisfy the condition of approximate Markov blanket. However, the FCBF considers only the relevance between the feature and the class when selecting the best features, and fails to take into consideration the interaction between features. In this paper, we propose an improved FCBF to overcome this shortcoming. We also perform a comparative study for the evaluation of the performance of the proposed method.
The remainder of the paper is organized as follows. The filter methods based on the information entropy are introduced in Section 2. In Section 3, the computational results of the performance are presented. Finally conclusions are mentioned in Section 4.
2. Methods
2.1 IG
The IG filter method, originally proposed by Quinlan [25], is one of the most common univariate methods of evaluation attributes. This filter method assesses features based on their information gain and considers a single feature at a time. The information entropy is employed as a measure to rank variables. The entropy of a class feature Y is defined as follows [23].
| (1) |
where P(Y) is the marginal probability density function for the random variable Y.
The value of IG for the attribute feature X is then given by
| (2) |
where H(Y/X) is the conditional entropy of Y given X.
The IG filter method first assigns an orderly classification of all features. A threshold value is then adopted to select a certain number of features based on the order obtained. As IG is a univariate approach that ignores the mutual information between attribute features, the computing time of the method is fast. However, if the attribute features are highly correlated, the IG filter method produces less accuracy.
2.2 FCBF
The FCBF filter method [23] is a multivariate approach that considers feature-class and the correlation of the attribute features, that is, feature-feature correlation. This filter method starts by selecting a set of features that is highly correlated with the class based on the following measure of SU(symmetrical uncertainty) [23].
| (3) |
The basic idea of FCBF constructs the features that are more relevant to the class Y and removes the redundant features by the property of approximate Markov blanket [23]. The pseudocode of FCBF is as follows.
2.3 mRMR
The mRMR filter method [24] is another multivariate algorithm for the feature selection. The basic idea of the mRMR is to construct attribute features that are maximally relevant to the class and also minimally redundant between the attributes. The criteria of maximum-relevance and minimum-redundancy are based on mutual information. The measure of mutual information is given by
| (4) |
Based on the mutual information, feature selection must find a feature set S with m features {xi}, which jointly have the maximum-relevance on the class Y. The problem being considered here has the following formulation.
| (5) |
Practically, if the number of features is very large, the criterion (5) is hard to implement. Therefore, Peng et al. [24] proposed an alternative criterion for maximum-relevance.
| (6) |
The above criterion approximates the maximum-relevance with the mean value of all mutual information between each feature xi and class Y.
Peng et al. [24] also proposed the following criterion for the minimum-redundancy.
| (8) |
Therefore, the criterion that combines the above two criteria is as follows.
| (9) |
In practice, Peng et al. [24] suggested the incremental search method to find the near-optimal features. The method optimize the following condition.
| (10) |
The above problem is to find the m th feature from the set {X − Sm−1}
The pseudo-code for the mRMR algorithm is as follows.
2.4 I-FCBF(Improved FCBF)
In this section, we propose an improved FCBF by hybridizing mRMR and FCBF. The FCBF has the advantage of reducing the computational burden by eliminating the redundant features that satisfy the condition of approximate Markov blanket. However, FCBF considers only the relevance between the feature and class in order to select the best features. It fails to take into consideration the interaction between features. To overcome this shortcoming of FCBF, we incorporate FCBF into mRMR to select the relevant features. In other words, we adopt the criterion of (10) to consider the interaction between features. After the feature is selected, we exploit the same reduction technique using the approximate Markov blanket as in the FCBF.
The detailed procedure of the I-FCBF is as follows.
3. Computational Results
To evaluate the performance of the filter methods (IG, FCBF, mRMR, and I-FCBF), nine data sets were selected from literatures. For evaluating the classification accuracy, Ambroise and McLachlan [26] recommended the use of 10-fold cross validation. Therefore, the 10-fold cross validation for all data sets was adopted in our experiments. Accuracy results were obtained by varying the number of best features from 5 to 30. In the tables, the bold number denotes the best accuracy among the four filter methods.
The accuracy of the classifier can be described in terms of true positives (TP), true negatives (TN), false negatives (FN), and false positives (FP) such that:
| (11) |
In our experiments, the Gaussian radial kernel was employed for the classification performance of SVM. Additional parameters of SVM were used in the default values of R-code.
3.1 Biological data sets
The three datasets are shown in Table 1. The dataset Lymphoma [27] contained 4026 features, 96 samples, and 9 classes. The quantities of genes and samples in the NCI [28] data set were 9712 and 60, respectively. The target class has 9 states. In the Breast cancer [29] data set, there are composed of 24481 features and 97 samples. Among these samples, 46 of which are from patients who had labeled as relapse, the rest 51 samples are from patients who remained healthy and regarded as non-relapse.
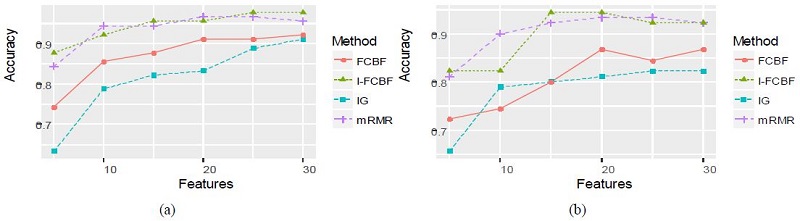
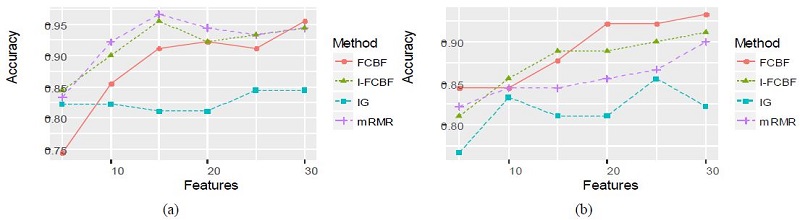
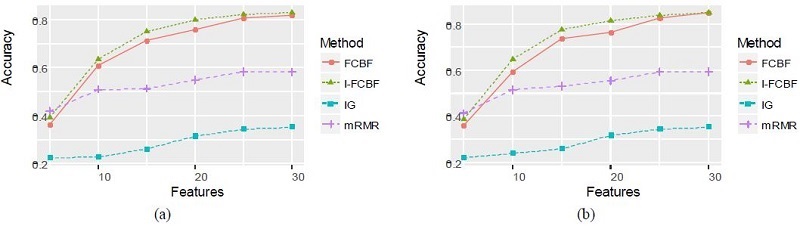
We compared our I-FCBF with three filter method based on the information entropy: IG, FCBF, mRMR. Tables 4 and 5 summarize the classification accuracy of NB and SVM, respectively, when using the four filetr methods. Table 4 shows that NB accuracy from using the IG method was the worst of four methods. The NB accuracy from using our IFCBF was better than the FCBF for most cases in the Breast cancer data set. For the Breast cancer data set, mRMR obtained relatively good results.
Table 5 for the SVM accuracy obtained nearly the same results as Table 4. The accuracy of the IG filter method was very poor, and the I-FCBF ontained better results than the FCBF for the Lyphoma and NCI data sets. However, for the Breast cancer data set, FCBF obtained better results than the I-FCBF. Tables 4 and 5 show that the NB and SVM accuracy produced the consistent results for the Lyphoma and NCI data sets. However, it can be seen that the results for the Breast cancer data set are highly dependent on the classifier.
The results of plotting the NB and SVM accuracy are shown in Figure 3 - 5.
3.2 Text data sets
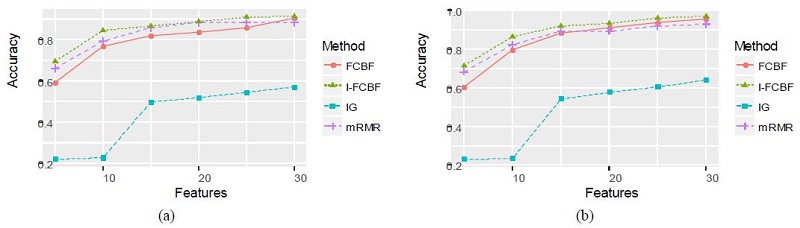
The characteristics of these data sets are shown in Table 2. These sample sizes are larger than the Biological data sets. The sample sizes of BASEHOCK, PCMAC, and RELATHE are 1993, 1943, and 1427, respectively. All of them are binary class data sets.
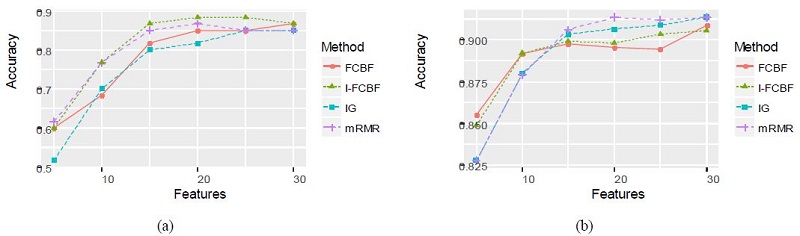
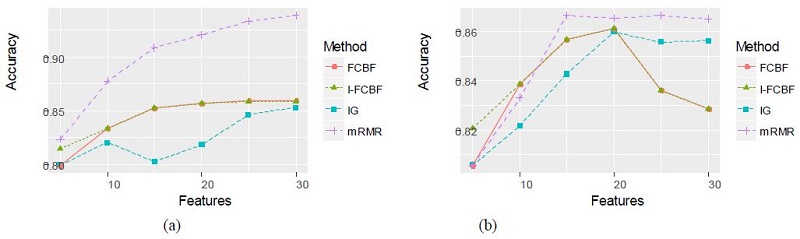
Tables 6 and 7 summarize the classification accuracy of the NB and SVM when using the four filter methods, respectively.
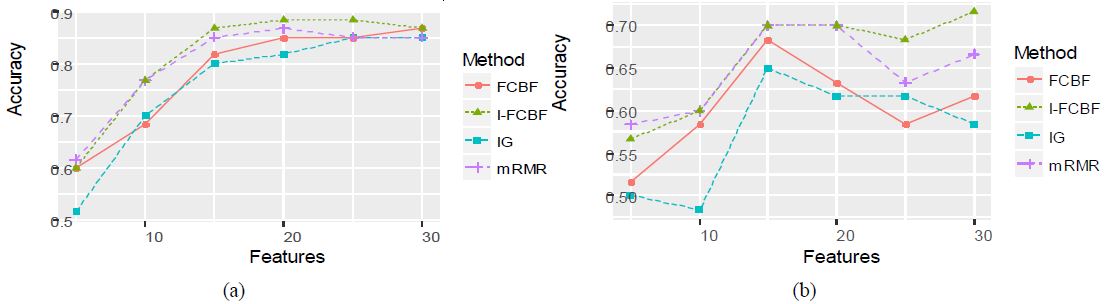
As can be seen, the NB and SVM accuracy of mRMR was the best among the four filter methods. In these cases, the IG method obtained relatively good results. As shown in Table 6 and 7, the accuracy of the IG method was better than that of the FCBF and I-FCBF for the case of 5 features. This implies that the reduction engine of FCBF using the approximate Markov blanket results in less efficiency for the selection of the best subset of the features. That is, it comes from the fulsome reduction of the promising subset of features, even although the approximate Markov blanket of the FCBF could reduce computational burden by removing redundant features. The results of plotting the NB and SVM accuracy are shown in Figure 6 - 8.
3.3 Multi-class data sets
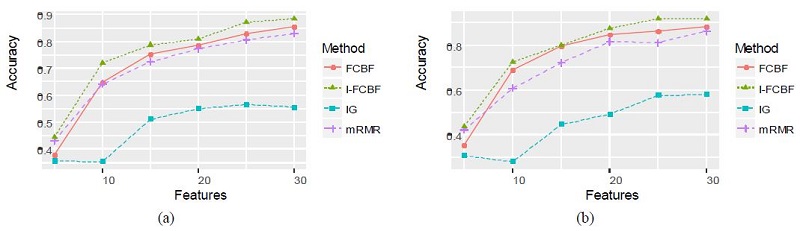
The three data sets are shown in Table 3, in which the class sizes are larger than those of two previous data sets. The class sizes of Isolet, COIL, and ORL are 26, 20, and 40, respectively.
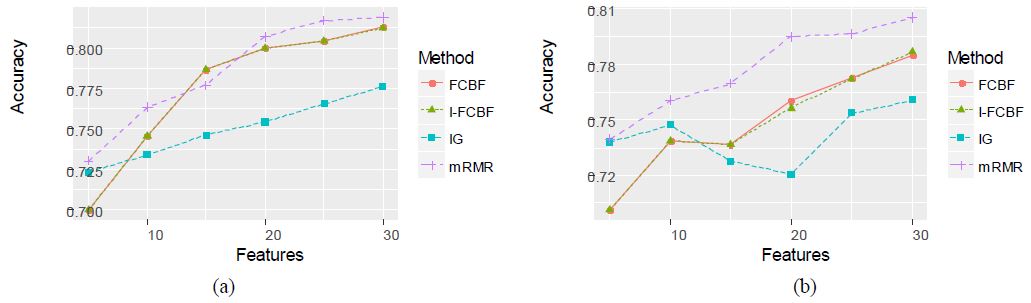
Tables 8 and 9 represent the classification accuracy of NB and SVM, respectively, when using the four filter methods. As shown in Tables 8 and 9, the NB and SVM accuracy of IFCBF was the best among the four filter methods. Unlike the data sets mentioned in Section 3.2, it can be seen that the reduction engine of I-FCBF does work well in constructing the best subset of features. That is, the approximate Markov blanket of the I-FCBF filter method seems to effectively remove the irrelevant and redundant features.
Specifically, the NB and SVM accuracy of the IG and mRMR was very poor for the Isolet data set. Remarkably, we noticed that for the case of 30 features, the SVM accuracy of mRMR and I-FCBF had a big gap between 0.5929 and 0.8500, respectively. For all multi-class data sets, the NB and SVM accuracy of our I-FCBF were also better than that of FCBF. The results of plotting the NB and SVM accuracy are shown in Figure 9 - 11.
4. Conclusions
Many feature selection methods have been developed to reduce the dimensionality of data sets. In this paper, we focused on the filter methods based on information entropy: IG, FCBF, and mRMR. The IG filter method is a univariate approach to evaluate the relevance of each feature individually, and a subset of features having the highest ranks is then selected. The IG method is computationally efficient. However, it produces a less accurate solution due to the ignorance of the dependency betwwen features. To overcome the shortcoming of the univariate method, multivariate algorithms have been proposed in the literature. FCBF and mRMR are well-known as efficient multivariate approaches.
The FCBF filter method has the advantage of reducing computational burden by removing irrelevant and redundant features that satisfy the condition of approximate Markov blanket. However, the FCBF considers only the relevance between the feature and class in order to select the best subset of features. It fails to consider the interaction between features. In this paper, we proposed an improved FCBF by hybridizing mRMR and FCBF. To overcome the shortcoming of FCBF, we incorporated FCBF into mRMR to select relevant features. In other words, we adopted the criterion of (10) to consider the interaction between features. After selecting the feature, we exploited the same reduction technique using the approximate Markov blanket as in FCBF.
We also performed a comparative study to evaluate the performance of the proposed method. We conducted experiments with three data sets from previous studies: biological, text, and multi-class data sets. We noticed that our I-FCBF obtained better results than the other methods for the biological and multi-class data sets. Remarkably, our I-FCBF filter method was performed the best for multi-class data sets with many classes.
However, for the text data sets with binary-class, our I-FCBF method failed to obtain the best results due to the fulsome reduction of the features using the approximate Markov blanket. In the next step, it needs to be developed a compensated and efficient reduction-engine to remove irrelevant and redundant features.
References
- M. Hall, “Correlation-based feature selection for machine learning”, PhD thesis, Citeseer, (1999).
- Z. Zhao, H. Liu, “Searching for interacting features”, International Joint Conference on Artificial Intelligence, 7, p1156-1161, (2007).
-
I. Guyon, J. Weston, S. Barnhill, and V. Vapnik, “Gene selection for cancer classification using support vector machines”, Machine Learning, 46, p389-422, (2002).
[https://doi.org/10.1023/A:1012487302797]

-
S. Maldonado, R. Weber, and J. Basak, “Simultaneous feature selection and classification using kernel-penalized support vector machines”, Information Sciences, 181(1), p115-128, (2011).
[https://doi.org/10.1016/j.ins.2010.08.047]

-
J. G. Bae, J. T. Kim, and J. H. Kim, “Subset selection in multiple linear regression: an improved tabu search”, Journal of Korean Society of Marine Engineering, 40(2), p138-145, (2016).
[https://doi.org/10.5916/jkosme.2016.40.2.138]

- I. Inza, B. Sierra, R. Blanco, and P. Larranaga, “Gene selection by sequential search wrapper approaches in microarray cancer class prediction”, Journal of Intelligent and Fuzzy Systems, 12(1), p25-33, (2002).
-
R. Ruiz, J. Riquelme, and J. Aguilar-Ruiz, “Incremental wrapper-based gene selection from microarray data for cancer classification”, Pattern Recognition, 39(12), p2383-2392, (2006).
[https://doi.org/10.1016/j.patcog.2005.11.001]

- S. Shreem, S. Abdullah, M. Nazri, and M. Alzaqebah, “Hybridizing ReliefF, mRMR filters and GA wrapper approaches for gene selection”, Journal of Theoretical and Applied Information Technology, 46(2), p1034-1039, (2012).
-
L. Chuang, C. Yang, K. Wu, and C. Yang, “A hybrid feature selection method for DNA microarray data”, Computers in Biology and Medicine, 41(4), p228-237, (2011).
[https://doi.org/10.1016/j.compbiomed.2011.02.004]

- W. Aiguo, A. Ning, C. Guilin, and L. Lian, “Hybridizing mRMR and harmony search for gene selection and classification of microarray data”, Journal of Computational Information Systems, 11(5), p1563-1570, (2015).
-
V. Vapnik, “Support-vector networks”, Machine Learning, 20, p273-297, (1995).
[https://doi.org/10.1007/BF00994018]

- J. Demsar, B. Zupan, M. W. Kattan, J. R. Beck, and I. Bratko, “Naive bayesian-based nomogram for prediction of prostate cancer recurrence”, Studies in Health Technology and Informatics, 68, p436-441, (1999).
-
H. Sun, “A naive Bayes classifier for prediction of multidrug resistance reversal activity on the basis of atom typing”, Journal of Medicinal Chemistry, 48(12), p4031-4039, (2005).
[https://doi.org/10.1021/jm050180t]

-
T. M. Cover, and P. E. Hart, “Nearest neighbor pattern classification”, IEEE Transactions on Information Theory, 13(1), p21-27, (1967).
[https://doi.org/10.1109/TIT.1967.1053964]

-
J. N. Morgan, and J. A. Sonquist, “Problems in the analysis of survey data, and a proposal”, Journal of the American Statistical Association, 58(302), p415-434, (1963).
[https://doi.org/10.1080/01621459.1963.10500855]

- J. A. Hartigrn, Clustering Algorithms, Wiley, New York, (1975).
-
L.E. Raileanu, and K. Stoffel, “Theoretical comparison between the Gini Index and information gain criteria”, Annals of Mathematics and Artificial Intelligence, 41(1), p77-93, (2004).
[https://doi.org/10.1023/B:AMAI.0000018580.96245.c6]

- M. Hall, and L. Smith, “Practical feature subset selection for machine learning”, Computer Science, 98, p181-191, (1998).
-
J. Yang, Y. Liu, Z. Liu, X. Zhu, and X. Zhang, “A new feature selection algorithm based on binomial hypothesis testing for spam filtering”, Knowledge-Based Systems, 24(6), p904-914, (2011).
[https://doi.org/10.1016/j.knosys.2011.04.006]

- Q. Gu, Z. Li, and J. Han, “Generalized fisher score for feature selection”, Proceedings of the International Conference on Uncertainty in Artificial Intelligence, (2011).
- X. He, D. Cai, and P. Niyogi, “Laplacian score for feature selection”, Advances in neural information processing systems, p507-514, (2005).
- K. Kira, and L. Rendell, “The feature selection problem: traditional methods and a new algorithm”, Proceedings of the Tenth National Conference on Artificial intelligence, AAAI Press, San Jose, CA, 2, p129-134, (1992).
- L. Yu, and H. Liu, “Feature selection for high-dimensional data: a fast correlation-based filter solution”, Proceedings of the Twentieth International Conference on Machine Learning, 3, p856-863, (2003).
-
H. Peng, F. Long, and C. Ding, “Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy”, IEEE Transactions on pattern analysis and machine intelligence, 27(8), p1226-1238, (2005).
[https://doi.org/10.1109/TPAMI.2005.159]

-
J. R. Quinlan, “Induction of decision trees”, Machine Learning, 1(1), p81-106, (1986).
[https://doi.org/10.1007/BF00116251]

-
C. Ambroise, and G. McLachlan, “Selection bias in gene extraction on the basis of microarray gene-expression data”, proceedings of the National Academy of Sciences, 99(10), p6562-6566, (2002).
[https://doi.org/10.1073/pnas.102102699]

-
A. A. Alizadeh, et al , “Distinct types of diffuse large B-cell lymphoma identitfied by gene expression profiling”, Nature, 403(6769), p503-511, (2000).
[https://doi.org/10.1038/35000501]

- U. Scherf, et al , “A cDNA microarray gene expression database for the molecular pharmacology of cancer”, 24(3), p236-244, (2000).
-
L. J. Vant’t Veer, et al , “Gene expression profiling predicts clinical outcome of breast cancer”, Nature, 415(6871), p530-536, (2002).
[https://doi.org/10.1038/415530a]